The CMI offers several orthogonal biophysical technologies to measure molecular interactions and to characterize molecular properties. To see how these technologies compare to each other see the Technologies Comparision Tables.
The CMI offers several orthogonal biophysical technologies to measure molecular interactions and to characterize molecular properties. To see how these technologies compare to each other see the Technologies Comparision Tables.
Bio-Layer Interferometry (BLI) is an optical technique for measuring macromolecular interactions by analyzing interference patterns of white light reflected from the surface of a biosensor tip. BLI experiments are used to determine the kinetics and affinity of molecular interactions. In a BLI experiment, one molecule is immobilized to a Dip and Read Biosensor and binding to a second molecule is measured. A change in the number of molecules bound to the end of the biosensor tip causes a shift in the interference pattern that is measured in real-time.
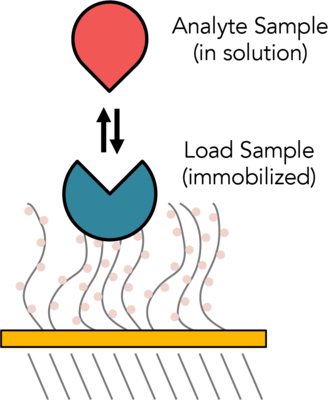

The CMI has two Sartorius (formerly ForteBio) instruments for measuring BLI: the Octet RED384 (now called Octet RH16) and the BLItz (now called Octet N1). The Octet RED384 is more sensitive and higher throughput and can be used for macromolecular and small molecule binding experiments. The BLItz uses a very small sample volume, but is more time consuming and cannot measure molecules smaller than 10 KDa.
Surface Plasmon Resonance (SPR) is an optical technique used to measure molecular interactions in real time. SPR can occur when plane-polarized light hits a metal film under total internal reflection conditions. SPR signal is directly dependent on the refractive index of the medium on the sensor chip. The binding of biomolecules results in changes in the refractive index on the sensor surface. In an SPR experiment, one molecule (the Ligand) is immobilized on a sensor chip and binding to a second molecule (the Analyte) is measured under flow. Response is measured in resonance units (RU) and is proportional to the mass on the surface, and for any given interactant, the response is proportional to the number of molecules bound to the surface. Response is recorded and displayed as a sensogram in real time. SPR experiments can be used to measure kinetic binding constants (ka, kd) and equilibrium binding constants (affinity, Ka = 1/Kd).
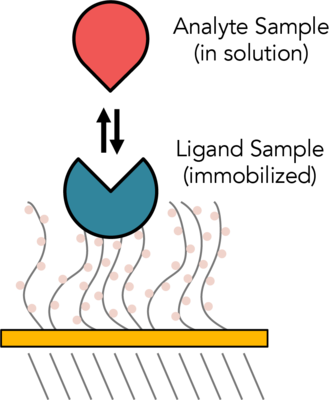

The CMI has a Biacore T200 from Cytiva, formerly GE Life Sciences.
MicroScale Thermophoresis (MST) is an immobilization-free technology for measuring biomolecular interactions. The MST instrument detects the motion of fluorescent molecules along a microscopic temperature gradient, which reflects changes in the molecular hydration shell, charge, or size. Since one or all of these parameters will change with virtually every binding event, a wide range of biomolecules can be measured, from ions and small molecule fragments to large macromolecular complexes, in small volumes (~20 μl), in a wide range of standard buffers and complex mixtures such as liposomes, detergent, serum, and cell lysates.
The CMI has a Monolith NT.115pico from NanoTemper Technologies.
Isothermal Titration Calorimetry (ITC) is a label-free method for measuring binding of any two molecules that release or absorb heat upon binding. ITC can be used to measure the thermodynamic parameters of biomolecular interactions, including affinity (KA), enthalpy (ΔH), entropy (ΔS), and stoichiometry (n). Energetically favorable binding reactions have negative free energy values, ΔG = RTlnKD. ΔG, has two energetic components, enthalpy (ΔH) and entropy (ΔS) and their contributions are expressed as: ΔG = ΔH -TΔS. In an ITC experiment, ΔH of binding is measured directly. The microcalorimeter has two cells: one contains water and acts as a reference cell, the other contains the sample, into which a binding partner is titrated using an injection syringe. Heat sensing devices detect temperature differences between the cells when binding occurs in the sample cell and give feedback to the heaters, which compensate for this difference and return the cells to equal temperature.
Heats measured in an ITC are a combination of heats of binding and heats of dilution, so a control experiment to measure heats of binding must be performed. Therefore, one experiment includes two titrations:
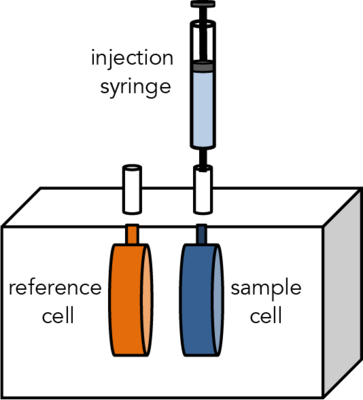
The CMI has a Microcal ITC200 from Malvern.
Differential Scanning Fluorimetry (DSF) measures protein unfolding by monitory changes in fluorescence as a function of temperature. Conventional DSF uses a hydrophobic fluorescent dye that binds to proteins as they unfold. NanoDSF measures changes in intrinsic protein fluorescence as proteins unfold.
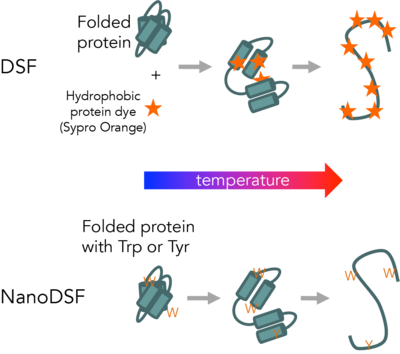
The CMI has a modified Life Technologies Quant Studio 6/7, for conventional DSF. The CMI has the Life Technologies Protein Thermal Shift Analysis software for DSF data fitting.
The CMI has a Prometheus NT.Plex instrument from NanoTemper Technologies with aggregation optics. The CMI has these Data collection and analysis software packages: PR.ThermControl for thermal stability data collection, PR.ChemControl for chemical stability data collection, PR.TimeControl for time interval data collection, and PR.Stability Analysis for advanced data analysis.
Circular Dichroism (CD) Spectroscopy is used to determine the optical isomerism and secondary structure of molecules. Circular dichroism (measured in molar ellipticity) is the difference in absorption of left-handed and right-handed circularly polarized light and can be observed in optically active molecules with chiral centers. Proteins have many chiral centers. CD spectra in the Far-UV region (185 – 250 nm) can be used to determine protein secondary structure. Characteristic peaks for Thermal stability (Tm) can be measured by following changes in molar ellipticity with increasing temperature.
The CMI has a Jasco J-1500 CD Spectropolarimeter with a Peltier temperature controller and single cuvette holder.
In April 2023, the Jasco J-815 was retired, after serving the BCMP department and HMS for almost 2 decades.
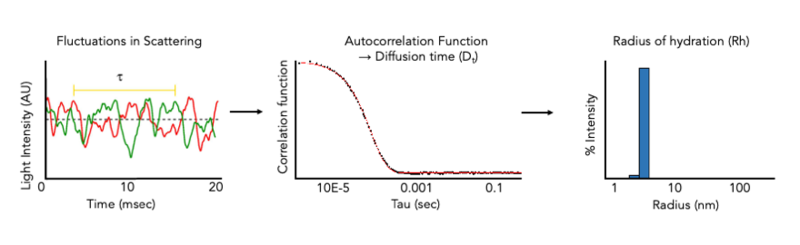
The CMI has Wyatt DynaPro Plate Reader III. The DynaPro Plate Reader III measures dynamic light scattering (DLS) and Static Light Scattering (SLS) at high-throughput on many sample types in standard microwell plates. and can be used to rapidly assess protein quality in solution. DLS measurements of hydrodynamic radius allow the monodispersity and aggregation state of a sample in solution to be measured in minutes and can be used to monitor protein quality during any (or all) stages of a protein purification project, or after storage. DLS can also be used to size a variety of nanoparticles, including liposomes, micelles, bicelles, and nanodiscs and for aggegation analysis of chemical compounds.

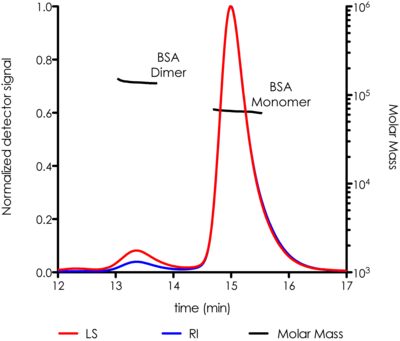
The CMI has a SEC-MALS system with a Wyatt Dawn Heleos Multi-Angle Light Scattering (MALS, 18-angle) Detector, with in-line DLS detector and Optilab TrEX refractive index detector. SEC-MALS is used to measure the weight-averaged molar mass of proteins (and other macromolecules) in solution, and can determine oligomeric state and sample polydispersity. Size-exclusion chromatography (SEC) separates molecules based on hydrodynamic volume, but is dependent on similarity to a set of reference standards for accurate mass determination and fails for elongated or sticky proteins. Multi-Angle Light static Scattering (MALS) is used to measure light scattering intensity accurately, which is proportional to the weight-averaged mass in solution. Combining SEC, MALS and concentration detectors in an SEC-MALS experiment allows for more accurate mass measurements that SEC or MALS alone.
By using two concentration detectors (RI and UV), the molar mass and weight-fraction of a modifier can be determined. This can be used to measure protein and modifier masses of integral membrane proteins in detergent micelles and of glycosylated proteins.
. 
Mass Photometry (MP) measures light scattering of single particles as they adsorb onto a glass microscope slide, and is used to rapidly determine mass, oligomeric state, and heterogeneity of a wide range of macromolecules and their complexes in solution, under equilibrium conditions and without the need for labels. Mass photometry can be used to determine masses of diverse macromolecules from 30 KDa to 5 MDa, including proteins, nucleic acids, lipids and small viruses, such as AAV.
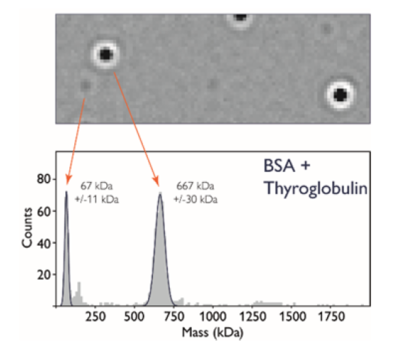 .
. 
The CMI has a Refeyn TwoMP mass photometry instrument from Refeyn, Ltd, with Accurion vibration-isolation bench.