For information on access fees, policies and getting started at the CMI, see the CMI Access Page.
MicroScale Thermophoresis (MST)
MST at the CMI
MicroScale Thermophoresis (MST) is an immobilization-free technology for measuring biomolecular interactions. The MST instrument detects the motion of fluorescent molecules along a microscopic temperature gradient, which reflects changes in the molecular hydration shell, charge, or size. Since one or all of these parameters will change with virtually every binding event, a wide range of biomolecules can be measured, from ions and small molecule fragments to large macromolecular complexes, in small volumes (~20 μl), in a wide range of standard buffers and complex mixtures such as liposomes, detergent, serum, and cell lysates.
The CMI has a Monolith NT.115pico from NanoTemper Technologies.
Monolith NT.115pico Instrument Detectors
Pico RED detector
- excitation wavelength: 600-650 nm
- eg. AlexaFluor647, NT647, Cy5
- fluorophore concentration ≥ 50 pM
- Kd range: pM - mM
Nano BLUE detector
- excitation wavelength 460-490 nm
- eg. fluorescein, AlexaFluor488, NT495, GFP
- fluorphore concentration ≥ 5 nM
- Kd range: nM - mM
Key Features
- fast measurement: Kd in about 10 min
- wide Kd range from pM/nM to mM range
- low sample consumption: sample volume (<10 µl per concentration)
- immobilization free, in-solution measurements
- measurements in complex mixtures (cell lysates, serum, detergents, liposomes)
- wide size range for interactants (from ions to MDa complexes)
MST Theory
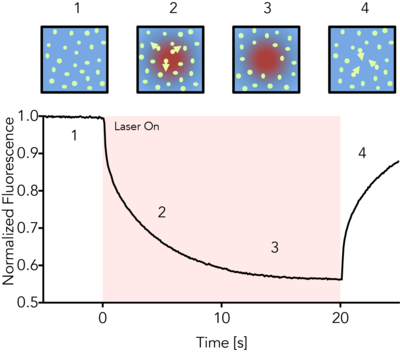
MicroScale Thermophoresis (MST) detects the motion of fluorescent molecules along a microscopic temperature gradient, which reflects changes in the molecular hydration shell, charge or size. A local temperature difference ΔT, induced by an infrared laser, leads to a local change in molecule concentration (depletion or enrichment), quantified by the Soret coefficient ST, chot/ccold=exp(-STΔT). This directed movement of molecules along a temperature gradient is termed "thermophoresis". Changes in the thermophoresis of a fixed concentration of fluorescent molecules in a constant buffer reflect changes in the size, charge or solvation entropy of the fluorescent molecules due to binding of the non-fluorescent partner (the titrant). Measuring this behavior at different concentrations of titrant allow quantification of the binding and determination of affinity.
Data Files - About CMI Data Files
Users are responsible for storage of all raw and processed data collected at the CMI.
- Users should have a plan to copy or transfer all raw and process data to their own local or cloud storage system.
- While the CMI allows temporary local storage of CMI User data on the instrument computer, we make no guarantees on the security or long-term availability of any data at the CMI.
- For most (but not all) CMI technologies, the raw data files and recommended readable exports are relatively small and can be readily transferred electronically.
- See specific instruments for exceptions and for details about the software, data file types and recommended data exports.
Data Sharing:
- Currently, a Generalist Repository is the recommended data repository for most CMI data types, as stable specialist data repositories have not been established.
Data Files - MST - Monolith NT.115pico
| Technology | Microscale Thermophoresis (MST) | ||
| Instrument | Nanotemper Monolith NT.115pico | ||
| Recommended Repository | Generalist Repository | ||
| Software Type | Data Collection | ||
| Current Version | MO.Control, Version 1.6.1 | ||
| Data Files (Type, ~size) | experiment file | .moc | 1-5 MB/experiment |
| raw data | .xslx | 10-25 MB/file | |
| Software Type | Data Analysis | ||
| Current Version | MO.Affinity Analysis, Version 2.3 | ||
| Data Files (Type, ~size) | analysis file | .nta | 1 MB/experiment |
| Readable Exports | processed data | .xslx | 10-25 KB/file |
| pdf report | 1 MB/report | ||
CMI Monolith MST Getting Started Guide
MicroScale Thermophoresis technology, from NanoTemper Technologies
Monolith NT.115 instrument product page, from NanoTemper Technologies
Required Supplies
- fluorescent target sample and non-fluorescent ligand sample and matched buffer
- MST capillaries (see below)
- 0.2 ml tubes for sample preparation (provided by CMI)
- pipetors and tips for liquid handing
NanoTemper Supplies
MST Capillaries
- Monolith NT.115 Standard Treated Capillaries, MO-K022 (available from the CMI, at cost)
- Monolith NT.115 MST Premium Coated Capillaries, MO-K025 (available from the CMI, at cost)
Labeling Kits (optional)
- NanoTemper Protein Labeling Kit RED-NHS 2nd Generation (Amine Reactive), MO-L011
- NanoTemper Protein Labeling Kit RED-MALEIMIDE 2nd Generation (Cys Reactive). MO-L014
- NanoTemper His-Tag Labeling Kit RED-tris-NTA 2nd Generation, MO-L018
Assay Buffers
- Many buffers are compatible with MST. It’s usually a good idea to start with a buffer system in which your proteins are well behaved.
- Addition of 0.05% Tween 20 or other detergent is usually required to prevent sticking of proteins to the capillaries.
- Each capillary should be prepared with identically matched buffer.
- Assay buffer (with detergent) is used to dilute the fluorescent molecule to 2X.
- Ligand buffer is used to dilute the ligand and should match the highest concentration of ligand
- 0.5-1 mg/ml BSA can also be used to minimize non-specific binding.
- Buffer cannot be opaque.
- High viscosity samples may be hard to fill (up to 10% glycerol is fine).
Samples
- All MST experiments are setup with one fluorescently-labeled molecule (the Target) at a fixed concentration mixed with various concentrations of a non-fluorescent molecule (the Ligand).
-
Concentration should be accurately measured
- Errors in Target concentration can affect fluorescent signal and may affect the fit
- Errors in the Ligand concentration will directly translate to errors in the KD
-
Protein aggregates will interfere with MST
- Filter or centrifuge samples before use.
- Assess protein heterogeneity via light scattering.
- Purify protein samples with soluble aggregates by size-exclusion chromatography.
Target Sample (the fluorescent molecule)
- ~200 µL/titration at >2X working concentration
- 5-20 µM unlabeled protein, if using a chemical labeling kit
-
RED detector:
- Stock concentration of labeled Target: > 10 nM
- Recommended working concentration: 5 nM (for KD > nM)
- Minimal working concentration: ≥ 50 pM (used for KD in pM range)
-
BLUE detector:
- Stock concentration of labeled Target: > 20 nM
- Recommended working concentration: 20 nM
- Minimal working concentration: ≥ 5 nM
Ligand Sample (the non-fluorescent binding partner)
- ~ 20 µL/titration, at 2X working concentration (bring the highest stock concentration available for an unknown KD)
- Recommended stock concentration ≥ 100X the expected KD
- Recommended working concentration ≥ 50X KD
