Mass Photometry (MP)
MP at the CMI
Mass Photometry (MP) measures light scattering of single particles as they adsorb onto a glass microscope slide, and is used to rapidly determine mass, oligomeric state, and heterogeneity of a wide range of macromolecules and their complexes in solution, under equilibrium conditions and without the need for labels. Mass photometry can be used to determine masses of diverse macromolecules from 30 KDa to 5 MDa, including proteins, nucleic acids, lipids and small viruses, such as AAV.
 .
. 
The CMI has a Refeyn TwoMP mass photometry instrument from Refeyn, Ltd, with Accurion vibration-isolation bench.
Key Features and Applications
Applications
-
Mass range: 30 kDa to 5000 kDa
- Proteins/complexes
- Nucleotides of up to ~5000 bp
- Empty vs Full ratio for AAV and small viruses
- Oligomeric state and heterogeneity
- Equilibrium dissociation constant (KD), for high affinity binders (~nM)
Key Features
- Small amount of sample required: typically a few μl at ~100 nM, measured at 10-20 nM
- Single particle counting
- Fast and easy to use
Mass Photometry Theory and Design
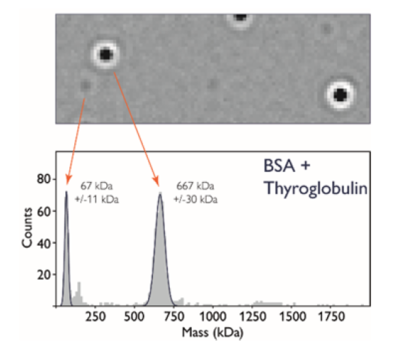
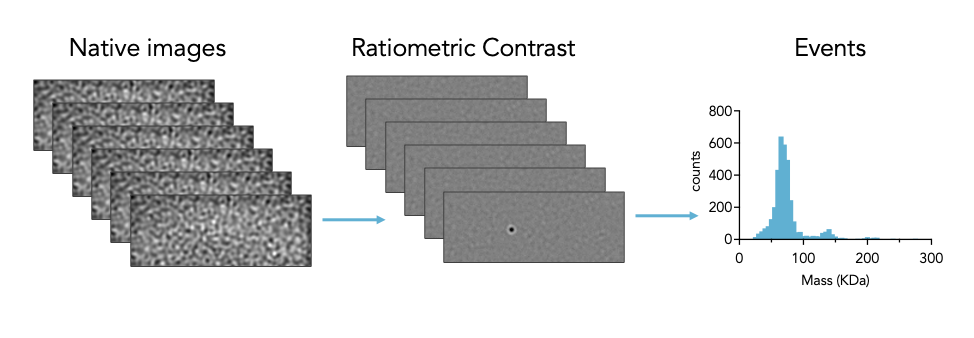
Built on the principles of interferometric scattering microscopy, Mass photometry measures the interference between the light scattered by a molecule in contact with the measurement surface and the light reflected by the surface. Data is collected as a short movie (1 minute) and then processed using ratiometric imaging, allowing weakly scattering single macromolecules to be distinguished from the high background signal. For scattering particles greater than 30 KDa and smaller than 100 nm, the point spread function representing each molecule that touches the surface has a signal intensity that is directly proportional to mass and refractive index. The instrument is calibrated using molecular standards of known mass with refractive index similar to the analyte of interest (e.g. protein, RNA, or DNA). Calculated masses of single particles are plotted as a histogram and can be counted or fit to Gaussian distribution.
Data Files - About CMI Data Files
Users are responsible for storage of all raw and processed data collected at the CMI.
- Users should have a plan to copy or transfer all raw and process data to their own local or cloud storage system.
- While the CMI allows temporary local storage of CMI User data on the instrument computer, we make no guarantees on the security or long-term availability of any data at the CMI.
- For most (but not all) CMI technologies, the raw data files and recommended readable exports are relatively small and can be readily transferred electronically.
- See specific instruments for exceptions and for details about the software, data file types and recommended data exports.
Data Sharing:
- Currently, a Generalist Repository is the recommended data repository for most CMI data types, as stable specialist data repositories have not been established.
Data Files - Mass Photometry - Refeyn TwoMP
Due to the large filesize of Mass Photometry data, the CMI instrument computer will be regularly purged of older data. Users should take extra care after each experiment to store their raw and processed data files in their own storage system.
|
Technology |
Mass Photometry | ||
| Instrument | Refeyn TwoMP | ||
| Recommended Repository | Generalist Repository | ||
| Software Type | Data Collection | ||
| Current Version | AccquireMP, version 2024 R1.1 | ||
| Data Files (Type, ~size) | processed data file (regular) | .mpr | ~30 MB/measurement |
| processed data file (large) | .mpr | ~125 MB/measurement | |
| Software Type | Data Analysis | ||
| Current Version | DiscoverMP, version 2024 R1 | ||
| Data Files (Type, ~size) | processed results (regular) | .mpr | ~30 MB/measurement |
| processed results (large) | .mpr | ~125 MB/measurement | |
| mass calibration | .mc | ~1 KB/measurement | |
| workspace file | .dmp | ~500 bytes/experiment | |
| Readable Exports | histogram | .png | ~200 KB/measurement |
| events file | .csv | ~0.5-2 MB/measurement | |
CMI Version History:
V2023-R2 September 2023
V2023-R1 March 2023
V2022-R1 Initial Version
Light Scattering Data Collection Services
Light Scattering Services Description
In addition to instrument training, the CMI is now offering basic Light Scattering Data Collection services, including:
Static Light Scattering: SEC-MALS or Mass Photometry
- mass determination in solution
- oligomeric state
Dynamic Light Scattering
- hydrodynamic radius
- buffer optimization
Mass Photometry Service Overview
Mass Photometry
Standard Mass Photometry Service
For proteins and protein complexes at least 30 KDa, without high molecular weight modifiers (modifiers of mass >5% total).
- Detector calibration with your buffer using a protein standard
-
Basic Data Analysis, in triplicate,
- Mass for each major peak
- Population distribution of major peaks
AAV Mass Photometry Services
For AAVs (replication-incompetent, ~3-5 MDa). Requires Empty Capsid control, provided by the user.
Empty/Full Service
- Detector calibration with your buffer using a protein standard
-
Basic AAV Analysis, in triplicate,
- Empty/Full population distribution
AAV Extended Service
- Detector calibration with your buffer using a protein standard and a DNA standard on poly-lysine coated slides
-
Extended AAV Analysis, in triplicate,
- Empty/Full population distribution
- Mass calculated for DNA cargo
SEC-MALS Service Overview
Size-exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS)
CMI SEC-MALS System: Wyatt Dawn Heleos with in-line DLS, RI UV, Agilent chromatography
Standard SEC-MALS Service
for proteins and protein complexes between 15 KDa and 1 MDa. Prior purification by size-exclusion chromatography (SEC) is strongly recommended.
Includes:
- Column equilibration with your buffer (pH at or below pH 7.5) or with PBS
- BSA Standard to test column performance and normalize detectors
- SEC separation of your protein sample(s)
-
Basic Data analysis, including:
- Molar Mass for each major peak
- Hydrodynamic Radius from DLS (when light scattering signal is sufficient)
Additional Services
-
Protein Conjugate Analysis
- required for samples with high molecular weight modifiers, such as glycosylation or detergent micelle (modifiers of mass >5% total)
- requires modifier dn/dc
- dn/dc measurements of detergent or protein samples
DLS Service Overview
Dynamic Light Scattering (DLS)
CMI DLS Instrument: Wyatt Dyanpro Plate Reader III
Standard DLS Service
For proteins and biopolymers up to 1000 nm in diameter. Includes:
- Cumulants and Regularization fit of Hydrodynamic Radius (Rh), 1 nm - 1000 nm
- Polydispersity Analysis
Additional Services
- Aggregation Analysis - Small molecule
- Buffer optimization
Data Collection Fees Summary
Data Collection
- Limited Data Collection Services are offered.
- Service fees are based on labor and supplies costs, and will be charged for all completed services, regardless of experimental outcome.
-
Before submitting samples for data collection, users must approve the estimated charges and be given a date and time for sample delivery.
- External Users will also be required to submit a PO and a signed CMI User Agreement.
-
Most CMI Data Collection Services include a setup fee plus a per-sample data collection fee.
- Some services include replicate measurements by default in the per-sample fee. For others, there is a reduced-price replicate measurement fee, if collected in the same dataset.
- Nanobody services not available to commercial users at this time.
- Current Harvard Life Lab commercial users are offered a 25% discount off the standard commercial rates.
Assay Buffers
- Buffers should be filtered to remove dust and particulates which will scatter light.
- Mass photometry is compatible with a range of buffers, but should avoid scattering particles when possible, including carrier proteins and detergents.
-
Detergents will cause background and at concentrations above the CMC, detergent micelles will scatter light. This makes measurement of membrane proteins very difficult.
- Use the lowest concentration of detergent possible, preferably below the CMC.
- Under limited circumstances, membrane proteins in detergent have been analyzed by mass photometry, but scattering of the empty micelles may complicate interpretation.
Samples
-
The ideal concentration range for sample measurement is 10-20nM. Samples should be prepared at 100-200nM for best results, as you will typically perform an ~8-10x dilution when mixing your sample into the buffer droplet.
-
The Concentration Calculator in the AcquireMP Tools menu can help with this.
-
-
You may need to try multiple concentrations to find the optimal conditions for your protein.
-
Calibration standards should be measured in sample assay buffer.
Calibration Standards
-
Calibrate mass at least every 3 hours.
-
Calibrate in assay/sample buffer, as the refractive index of the solvent impacts the ratiometric contrast.
-
Mass standards must have a similar refractive index to your sample particles.
-
Use protein standards for protein samples, a DNA standard or ladder for DNA samples, etc.
-
-
The CMI provides a 1000X Protein Calibration Mix (10µL aliquots of 3µM Thyroglobulin 10µM BSA in PBS and 5% glycerol) stored in 10µL aliquots in the -20˚C freezer below the instrument. Perform two 1:10 dilutions of the calibration mix with your buffer. The first dilution (1:10) can be stored at 8˚C for ~7 days. The second dilution (1:100) is the working stock which you will dilute into the buffer droplet.
-
For, specific instructions on preparing DNA and RNA calibrants see the CMI’s Guide to Measuring Nucleic Acids.
Provided by the CMI:
- Reusable Sample Well Cassettes (6-well silicone gaskets)
- Immersion oil: Zeiss Immersol 518 F
- Whatman® lens cleaning tissue, Grade 105
- Isopropanol and ultra-pure water, for cleaning
- No. 1.5H high precision glass coverslips (24x50 mm), Thorlabs CG15KH
- 1000x Protein Calibration Mix (10µL aliquots of 3µM Thyroglobulin, 10µM BSA in PBS with 5% glycerol)
Purchased by the User:
- Optional Pre-cleaned Sample slides: Refeyn Sample Carrier Slides
-
Optional Supplies (varies by experiment):
- Poly-lysine: Sigma P4832 (for measuring nucleic acids)
-
Calibration standards for non-protein samples:
- Invitrogen Low DNA Mass Ladder: Invitrogen 10068013
- Millennium RNA Marker: ThermoFisher AM7150
- Empty AAVs, high mass protein standard
