Size exclusion chromatography with multi-angle static light scattering (SEC-MALS) is used to accurately measure weight-averaged masses (Mw) of macromolecules in solution by measure the intensity of scattered light of a sample as it elutes from an SEC column.
Size-Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS)
SEC-MALS Instrumentation
SEC-MALS at the CMI
The CMI has a SEC-MALS system with a Wyatt Dawn Heleos Multi-Angle Light Scattering (MALS, 18-angle) Detector, with in-line DLS detector and Optilab TrEX refractive index detector. SEC-MALS is used to measure the weight-averaged molar mass of proteins (and other macromolecules) in solution, and can determine oligomeric state and sample polydispersity. Size-exclusion chromatography (SEC) separates molecules based on hydrodynamic volume, but is dependent on similarity to a set of reference standards for accurate mass determination and fails for elongated or sticky proteins. Multi-Angle Light static Scattering (MALS) is used to measure light scattering intensity accurately, which is proportional to the weight-averaged mass in solution. Combining SEC, MALS and concentration detectors in an SEC-MALS experiment allows for more accurate mass measurements that SEC or MALS alone.
By using two concentration detectors (RI and UV), the molar mass and weight-fraction of a modifier can be determined. This can be used to measure protein and modifier masses of integral membrane proteins in detergent micelles and of glycosylated proteins.
. 
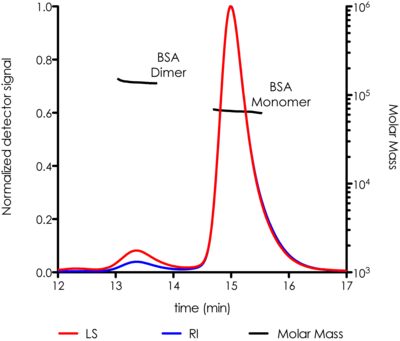
SEC-MALS System Components
MALS
- Wyatt Dawn Heleos II Multi-Angle Light Scattering (MALS) detector
- Wyatt Optilab T-rex Refractive Index Detector
- Wyatt In-line QELS detector for Dynamic Light Scattering
Chromatography
- Agilent 1260 Infinity Isocratic Liquid Chromatography System
- Agilent 1260 Infinity Autosampler
- inline solvent degasser
- variable wavelength UV detector
There is no fraction collector on this system, as it is intended for analytical and not preparative chromatography.
SEC-MALS Column Data
Data Files - About CMI Data Files
Users are responsible for storage of all raw and processed data collected at the CMI.
- Users should have a plan to copy or transfer all raw and process data to their own local or cloud storage system.
- While the CMI allows temporary local storage of CMI User data on the instrument computer, we make no guarantees on the security or long-term availability of any data at the CMI.
- For most (but not all) CMI technologies, the raw data files and recommended readable exports are relatively small and can be readily transferred electronically.
- See specific instruments for exceptions and for details about the software, data file types and recommended data exports.
Data Sharing:
- Currently, a Generalist Repository is the recommended data repository for most CMI data types, as stable specialist data repositories have not been established.
Data Files - SEC-MALS - Dawn Heleos II
| Technology | Size Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS) | ||
| Instrument |
Agilent 1260 Infinity LC System with variable UV detector Wyatt Dawn Heleos II MALS detector Wyatt Optilab T-rEX Refractive Index Detector |
||
| Recommended Repository | Generalist Repository | ||
| Software Type | Data Collection & Analysis | ||
| Current Version | Astra, Version 7. | ||
| Data Files (Type, ~size) | experiment file | .afe7 | 3-5 MB/measurement |
| Readable Exports | basic collection | .csv | 4 MB/measurement |
| report | 300-350 KB/measurement | ||
| easi graph | .csv | 5 MB/experiment | |
| easi table | .csv | 3-5 KB/experiment | |
Light Scattering Data Collection Services
Light Scattering Services Description
In addition to instrument training, the CMI is now offering basic Light Scattering Data Collection services, including:
Static Light Scattering: SEC-MALS or Mass Photometry
- mass determination in solution
- oligomeric state
Dynamic Light Scattering
- hydrodynamic radius
- buffer optimization
Mass Photometry Service Overview
Mass Photometry
Standard Mass Photometry Service
For proteins and protein complexes at least 30 KDa, without high molecular weight modifiers (modifiers of mass >5% total).
- Detector calibration with your buffer using a protein standard
-
Basic Data Analysis, in triplicate,
- Mass for each major peak
- Population distribution of major peaks
AAV Mass Photometry Services
For AAVs (replication-incompetent, ~3-5 MDa). Requires Empty Capsid control, provided by the user.
Empty/Full Service
- Detector calibration with your buffer using a protein standard
-
Basic AAV Analysis, in triplicate,
- Empty/Full population distribution
AAV Extended Service
- Detector calibration with your buffer using a protein standard and a DNA standard on poly-lysine coated slides
-
Extended AAV Analysis, in triplicate,
- Empty/Full population distribution
- Mass calculated for DNA cargo
SEC-MALS Service Overview
Size-exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS)
CMI SEC-MALS System: Wyatt Dawn Heleos with in-line DLS, RI UV, Agilent chromatography
Standard SEC-MALS Service
for proteins and protein complexes between 15 KDa and 1 MDa. Prior purification by size-exclusion chromatography (SEC) is strongly recommended.
Includes:
- Column equilibration with your buffer (pH at or below pH 7.5) or with PBS
- BSA Standard to test column performance and normalize detectors
- SEC separation of your protein sample(s)
-
Basic Data analysis, including:
- Molar Mass for each major peak
- Hydrodynamic Radius from DLS (when light scattering signal is sufficient)
Additional Services
-
Protein Conjugate Analysis
- required for samples with high molecular weight modifiers, such as glycosylation or detergent micelle (modifiers of mass >5% total)
- requires modifier dn/dc
- dn/dc measurements of detergent or protein samples
DLS Service Overview
Dynamic Light Scattering (DLS)
CMI DLS Instrument: Wyatt Dyanpro Plate Reader III
Standard DLS Service
For proteins and biopolymers up to 1000 nm in diameter. Includes:
- Cumulants and Regularization fit of Hydrodynamic Radius (Rh), 1 nm - 1000 nm
- Polydispersity Analysis
Additional Services
- Aggregation Analysis - Small molecule
- Buffer optimization
Data Collection Fees Summary
Data Collection
- Limited Data Collection Services are offered.
- Service fees are based on labor and supplies costs, and will be charged for all completed services, regardless of experimental outcome.
-
Before submitting samples for data collection, users must approve the estimated charges and be given a date and time for sample delivery.
- External Users will also be required to submit a PO and a signed CMI User Agreement.
-
Most CMI Data Collection Services include a setup fee plus a per-sample data collection fee.
- Some services include replicate measurements by default in the per-sample fee. For others, there is a reduced-price replicate measurement fee, if collected in the same dataset.
- Nanobody services not available to commercial users at this time.
- Current Harvard Life Lab commercial users are offered a 25% discount off the standard commercial rates.
Generally, SEC-MALS data collection is done in ASTRA 7 with HPLC control.
CMI SEC-MALS Getting Started Guide
CMI SEC-MALS Guide to Protein Conjugate Analysis, guide to measuring dn/dc values and performing protein conjugate data analysis.
CMI SEC-MALS Guide to Working with Custom Solvents, for solvents that cannot use PBS or water as the solvent model.
SEC-MALS Technology page from Wyatt Technologies.
- purified protein samples (must be filtered before loading)
- running buffer (1L)
- analytical SEC column (the CMI usually has 1-2 working SEC-MALS columns that users may borrow, if their samples are purified by SEC and in an appropriate buffer)
Assay Buffers
- Running buffer should always be chosen to be compatible with both the SEC column and the protein sample.
- Recommended Buffer: 25 mM HEPES pH 7-7.5, 150 mM NaCl (filtered).
-
Make sure you know the buffer compatibility of the SEC column you are using.
- Most silica columns will not tolerate pH above 7.5.
- Some buffer components (e.g. glycerol) will require customization of the solvent profile. Take note of the absolute refractive index of the solvent after equilibration and before data collection.
Samples
-
The sample should be prepared in running buffer to minimize RI peak due to sample solvent.
- This is particularly important for samples that run near the solvent peak.
- It is good practice to run a size-exclusion chromatography purification prior to analytical SEC-MALS to buffer exchange and clear aggregates. This is a requirement if you are using a shared CMI column.
- Samples must be filtered or centrifuged prior to injection.
-
Concentration should be accurately measured to assess column recovery.
- Know the concentration (in mg/ml) of your protein.
- Know the UV extinction coefficient (in ml/mg•cm) of your protein
-
Protein aggregates can damage the column.
- Filter or centrifuge samples before use.
- Assess protein heterogeneity via dynamic light scattering (in the DynaPro plate reader).
- Purify protein samples with soluble aggregates by size-exclusion chromatography.
-
Recommended protein concentration varies depending on protein mass:
-
Scattering is proportional to mass
- Larger proteins require less sample than smaller proteins.
-
Typical range 5 – 500 µg/injection
- BSA (67 KDa) 100 µl at 2 mg/ml always gives a good light scattering signal
-
Scattering is proportional to mass
-
Sample volume:
- Maximum injection volume: 100 µl
- Minimum injection volume: 5 µl
-
Glass autosampler vial with low volume insert has 10 µl dead volume.
- Fill vial with at least 110 µl for a 100 µl injection.
